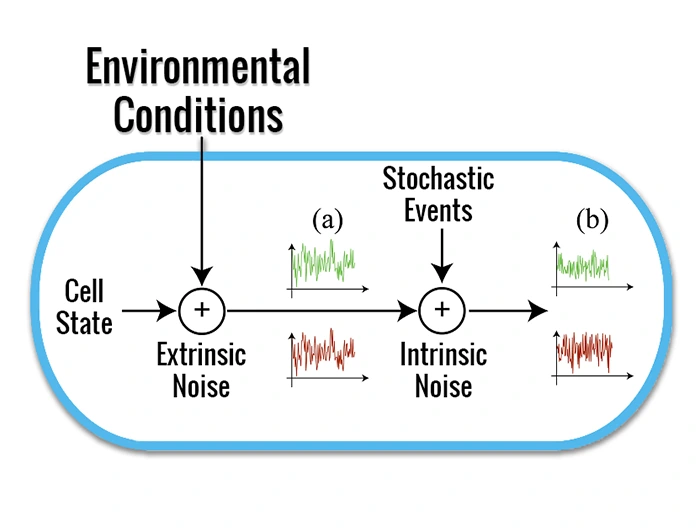
The stochasticity due to the infrequent collisions among low copy-number molecules within the crowded cellular compartment is a feature of living systems. Single cell variability in gene expression within an isogenic population (i.e. biological noise) is usually described as the sum of two independent components: intrinsic and extrinsic stochasticity. Intrinsic stochasticity arises from the random occurrence of events inherent to the gene expression process (e.g. the burst-like synthesis of mRNA and protein molecules). Extrinsic fluctuations reflect the state of the biological system and its interaction with the intra and extracellular environments (e.g. concentration of available polymerases, ribosomes, metabolites, and micro-environmental conditions). A better understanding of cellular noise would help synthetic biologists design gene circuits with well-defined functional properties.
Rif.
Bandiera L. et al. Phenotypic variability in synthetic biology applications: dealing with noise in microbial gene expression. Frontiers in Microbiology ISSN: 1664-302X (2016) 7:479 DOI: 10.3389/fmicb.2016.00479
Bandiera L. et al. Experimental measurement and mathematical modeling of biological noise arising from transcriptional and translational regulation of basic synthetic gene-circuits. Journal of Theoretical Biology ISSN: 0022-5193 (2016) 395:153-160 DOI: 10.1016/j.jtbi.2016.02.004