Contact details
+39 0577 585730
Università di Siena - Dip. di Biotecnologie Mediche,
Policlinico Le Scotte - Palazzina amministrativa, Viale Bracci, 2 - 53100 - Siena.
Unità Bioingegneria, Piano 1. Go to Google map
Simone Furini, PhD, is a researcher at the Department of Medical Biotechnologies of the University of Siena. His main area of expertise is the simulation of biological systems at the molecular and cellular level.
Research by Simone Furini
- Details
- Molecular Dynamics
When calculating the free-energy of ions permeating membrane proteins, an initial decision to make is how many ions needs to be included in the calculation. This decision is usually based on previous knowledge (both experiments and simulations), and on the intuition of the researcher. But intuition might be wrong… And taking the number of ions wrong might render free-energy profiles that are not r...
- Details
- Molecular Dynamics
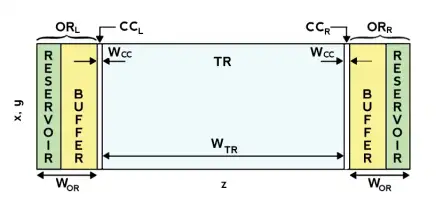
Periodic Boundary Conditions are almost universally adopted in simulations of biological systems at the microscopic scale, in order to replicate as close as possible the experimental environment, while keeping the number of particles to a minimum. While PBC are generally great, they do not allow to simulate gradients in-between the two sides of the simulation box, as these are actually the same si...
- Details
- Molecular Dynamics
Na+ channels have high-conductivity to sodium ions, while at the same time being almost completely impermeable to potassium ion. The first characteristic suggests loose atomic interactions between the permeating particles and the channel. On the other hand, it is hard to imagine how selectivity between two similar ion species might be possible under these loose atomic interactions. In this study,...
- Details
- Molecular Dynamics
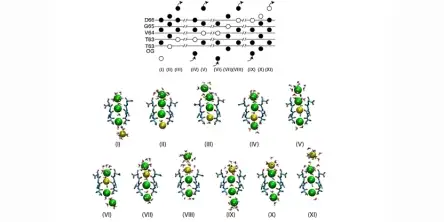
The selectivity of ion channels to different ion species is crucial for important biological processes, as nerve transmission and muscular contraction. Thanks to the availability of experimental atomic structures of several ion channels, including K+-channels, Na+-channels and Ca2+-channels, it is now possible to investigate the microscopic details of conduction and selectivity. As ion conduction...
- Details
- Molecular Dynamics
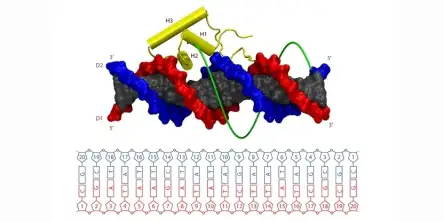
The binding of proteins to DNA controls many cellular events, including gene expression. In order to work properly, DNA binding proteins need to find their target sequences on DNA with high speed and accuracy. The recognition of the specific DNA binding site requires intimate contacts between the protein and the DNA molecule. However, strong atomic interactions between the two molecules are expect...
Latest by Simone Furini
- Free-energy profiles and number of permeation ions
- Concentration gradients in simulations with PBC
- Selectivity in bacterial Na+ channels
- Molecular Dynamics simulations reveal how K+-channels might conduct K+ ions at high speed
- Molecular Dynamics simulations can analyze how protein might move along DNA sequences