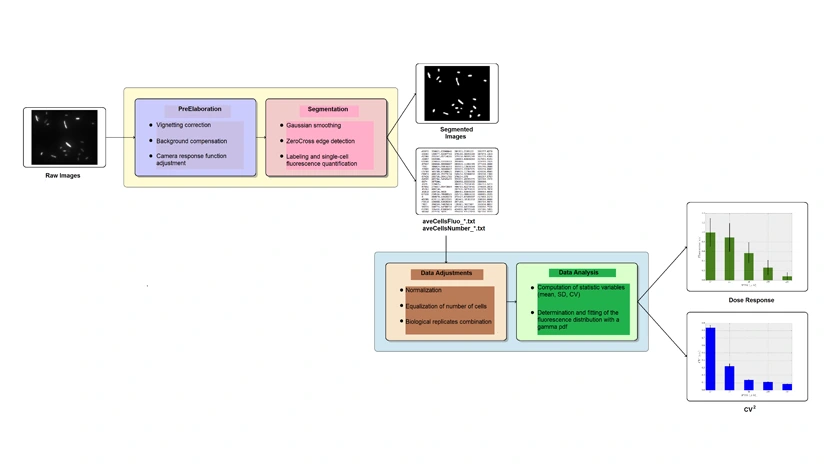
The evaluation of phenotypic variability within an isogenic population of bacterial cells, requires the quantification of gene expression at single cell level. Flow cytometers and microscopy set-ups equipped with microfluidic devices and incubation chambers are instruments capable of evaluating the level of specific proteins in individual cells. However they are expensive and generally unavailable in small laboratories. To promote the study of phenotypic noise, making it available to a larger base of researchers, we have developed MUSIQ (Microscope flUorescence SIngle cell Quantification), an open source software package that allows the quantification of the level of fluorescent protein markers through the analysis of images acquired using a standard optical microscopy set-up. This system has been completely validated through the comparison of its results with those of a flow cytometer and is freely available in the download section.
MUSIQ (Microscope flUorescence SIngle cell Quantification)